| PyPI: |  |
 |
| Help! |  |
General Instructions (for SOFCs)¶
Generic analysis¶
These instructions will walk through calculating the tortuosity for the two
example files Electrolyte and
Cathode. In general, the
low-level methods _geo_dist(),
_calc_interface(),
_calc_tort(), and
_calc_euc_x() do all the “heavy”
lifting, while modules like
tortuosity_from_labels_x() and
run_full_analysis_lsm_ysz() wrap these
methods into convenient higher-level interfaces.
Put the downloaded files into a directory accessible to Python, and fire up a Python session (Jupyter, notebook, etc.)
Import the module:
>>> import fibtortuosity as ft
The following code will use the higher order function
tortuosity_from_labels_x()to calculate the tortuosity in the x direction (perpendicular to the electrolyte boundary) The x direction function includes some additional code (compared to the y and z versions) that calculates the euclidean distance from the boundary of the bulk electrolyte, rather than simply from the edge of the volume.geo,euc,tort, anddescwill contain the results of this calculation afterwards:>>> geo, euc, tort, desc = ft.tortuosity_from_labels_x('example_electrolyte.tif', ... 'example_electrolyte_and_cathode.tif', ... 'LSM', ... units='nm', ... print_output=True, ... save_output=False) Starting calculation on LSM in x direction. Loaded electrolyte data in 0:00:00.257833 seconds. Loaded cathode data in 0:00:00.252949 seconds. ImageDescription is: b'BoundingBox 3231.15 6462.29 0 4100.38 0 4000\n' Bounding box dimensions are: (3231.14, 4100.38, 4000.00) nm Voxel dimensions are: (16.16, 20.50, 20.00) nm Starting geodesic calculation at: 2016-04-05 15:52:57.412840 Geodesic calculation took: 0:00:07.159744 Calculating zero distance interface took: 0:00:00.207048 Calculating euclidean distance took: 0:00:00.378937 Calculating tortuosity took: 0:00:00.295921 Total execution time was: 0:00:08.597359
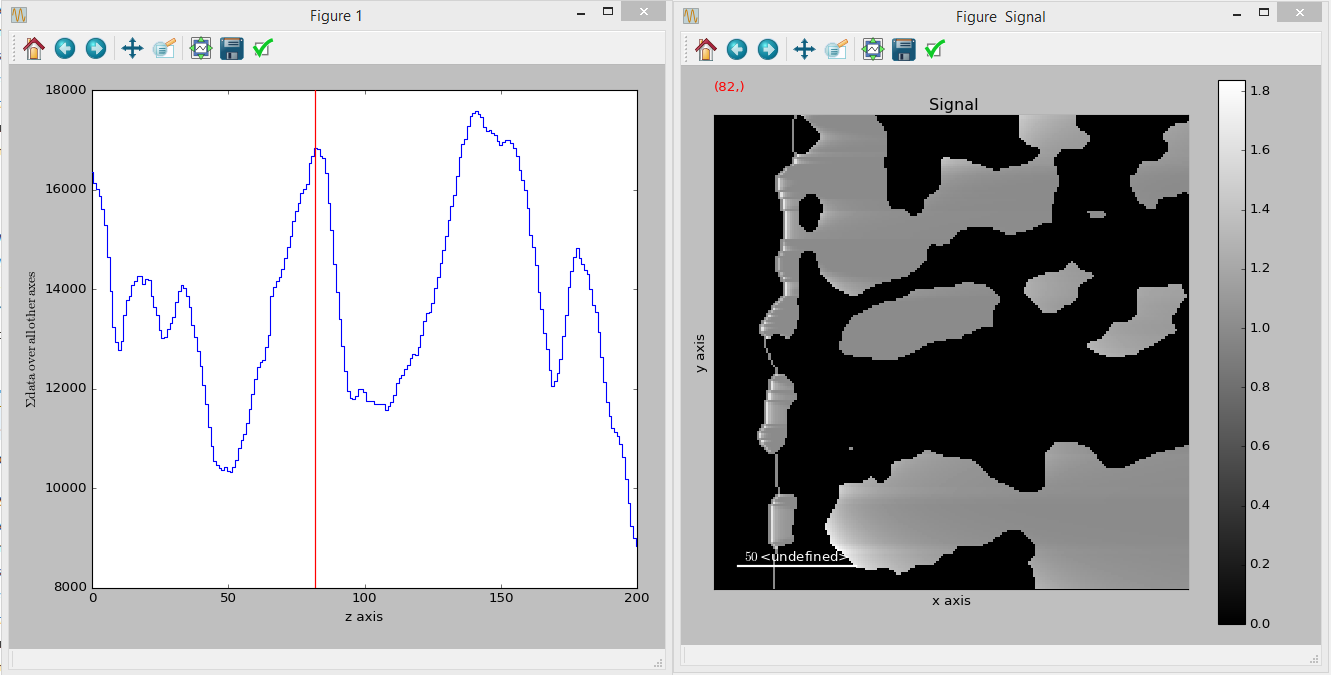
If HyperSpy is installed, it can be used to easily visualize the three-dimensional data that is produced as a result (example below is in Jupyter):
>>> %matplotlib qt4 >>> import hyperspy.api as hs >>> t_s = hs.signals.Image(tort) >>> for i, n in enumerate(['z', 'x', 'y']): ... t_s.axes_manager[i].name = n >>> t_s.plot()
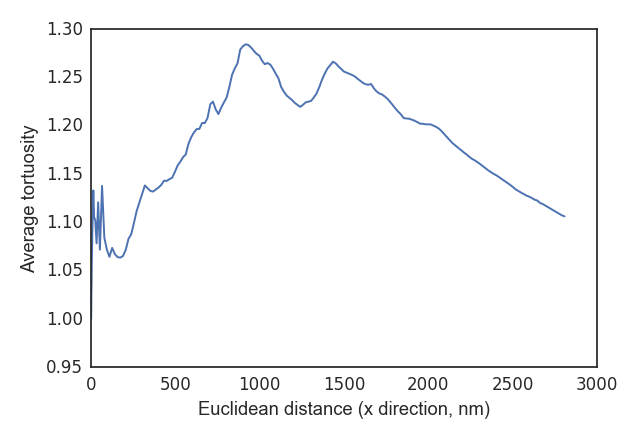
The
tortuosity_profile()andplot_tort_prof()methods can be used to visualize the average tortuosity over a dimension:>>> t_avg, e_avg = ft.tortuosity_profile(tort, euc, axis='x') >>> ft.plot_tort_prof(t_avg, e_avg, 'x')
To save the results, a variety of export options are available. The average profiles can be easily saved using
save_profile_to_csv(). The 3D tortuosity (or euclidean/geodesic distance) arrays can be saved as a 3D tiff using thesave_as_tiff()method. Also, if HyperSpy is being used, it can save the data in the.hdf5format (see its documentation for details). Furthermore, any of the Numpy methods for saving (such assave()orsavez()) can be used directly on the resulting arrays.To save profile:
>>> ft.save_profile_to_csv('profile.csv', e_avg, t_avg, 'x', 'LSM')
To save tiff file:
>>> # Using `desc` allows the file to be opened directly in Avizo >>> print(desc) b'BoundingBox 3231.15 6462.29 0 4100.38 0 4000\n' >>> ft.save_as_tiff('tortuosity.tif', tort, 'float32', desc) Writing TIFF records to tort_results.tif filling records: 100% done (12Mi+1003Ki+361 bytes/s) resized records: 31Mi+46Ki+716 bytes -> 8Mi+717Ki+565 bytes (compression: 3.59x)
This gives you the following output:
Tortuosity-profileandTortuosity-data
Full Analysis¶
In my personal research, I more fully combined these methods into the highest-level
function run_full_analysis_lsm_ysz(),
so I could just run one function, walk away, and let it run. It has
some additional useful features, like texting a number when it’s done or showing
a browser notification. Also, it’s written with specific materials in mind, so it
may take some tweaking to get it running for your specific needs, but I include
it here as an idea of what can be done with the underlying code.
To do a full analysis of a sample (all phases and all directions), I would run something like the following, making sure that each of the files specified are in the current directory:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | >>> import itertools
>>> import fibtortuosity as ft
>>> phases = ['LSM','YSZ','Pore']
>>> directions = ['x', 'y', 'z']
>>> for p, d in itertools.product(phases, directions):
... ft.run_full_analysis_lsm_ysz(electrolyte_file="bulkYSZ.tif",
... electrolyte_and_pore_file="bulkYSZandPRE.tif",
... electrolyte_and_lsm_file="bulkYSZandLSM.tif",
... electrolyte_and_ysz_file="bulkYSZandYSZ.tif",
... date='2016-04-05',
... phase=p,
... direction=d,
... npzfile=None,
... units='nm',
... delay=0,
... calculate_all=True,
... load_from_prev_run=False,
... create_hspy_sigs=False,
... save_avizo_tiff=True,
... tort_profile=True,
... save_tort_prof=True,
... in_ipython=False)
|
After this has run, the directory will contain all the tortuosity data saved
as .tif files, as well as all the average profiles for each phase and
each direction, which can then be plotted/analyzed however is necessary.